Chemicals that make bacteria lose their hair could be a new weapon in the fight against infections, and at the same time help to overcome the problem of antibiotic resistance
-
Peptide mimics stop bacterial hair growing, reducing their ability to stick to other cells and infect them
- Designer pilicides - clinical trials will determine efficacy
The ability of bacteria to cause diseases in humans or animals depends on several factors. One of these is so simple that it can be easily overlooked: bacteria need to stick to their host to be able to establish an infection. Many kinds of bacteria do this with the help of very thin hairs, or pili. For instance, the E. coli strains that cause urinary tract infections depend on pili, otherwise they would get washed away during urination.
Hair growth
Like our own hair and sheep wool, bacterial hairs are made of protein, albeit different kinds of protein, and they are a lot thinner than our hairs. The pili of E. coli are simple threads of protein subunits, which look like beads on a string.
Much like our own hair, pili grow as a result of new material feeding in at the root, so the tip is the oldest part of the pilus and remains unchanged. At the tip of E. coli pili we find the protein PapG, where Pap means 'pyelonephritis-associated pilus', after the type of urinary tract infection to which these pili were first connected. PapG is a sticky type of protein or 'adhesin'. Its task is to attach the bacterial cell to certain types of host cells, using the host's carbohydrate receptors.
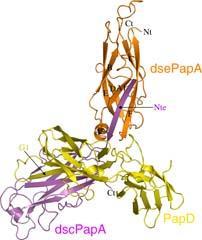
Following the pilus from the tip inward, we find a few other different Pap proteins, such as PapF, PapE, and PapK, and then a large number of copies of the main hair builder, PapA. In 2007, the team of Gabriel Waksman at University College London and Birkbeck College, London, in collaboration with Scott Hultgren at Washington University, St Louis, US, reported the crystal structure of PapA in an environment believed to reflect conditions in a real pilus (see Fig 1).1 As in most new crystal structures reported these days, the overall pattern in which the polypeptide chain winds it way through the three dimensional construct turned out to be a familiar one. In this case it was a variation of the immunoglobulin fold, a pattern of β-pleated sheets found in antibodies and many other proteins.
Intriguingly, the researchers found a β-strand missing on one side of the protein structure, and an unstruct-ured string of amino acids spare on the other side. The crystal structure also revealed that these two features latch into each other and hold the protein beads together, which are thus connected like the links of a chain, rather than beads on a string.
Chaperones zip in and out
It turns out that this remarkable connection relies on the work of other 'chaperone' and 'usher' proteins in the vicinity. The string of pilus proteins is assembled in the space (the periplasm) between the outer and inner membranes, a structural characteristic of Gram-negative bacteria. Because each building block has a gap in its structure and a spare length of polypeptide chain hanging around, these proteins are prone to unwanted interactions, which would lead to aggregation and loss of material. The 'molecular chaperones' protect these contact points from aggregation.
The protein PapD is a molecular chaperone that specialises in the assembly of pili and protection of their building blocks. This protein can bind to subunit proteins like PapA in a similar way to the interaction between PapA subunits. This enables a string of PapD sequence to latch into the PapA cavity.
The chaperone PapD escorts the building blocks towards the usher protein, PapC, which forms a pore through which the growing hair is threaded to the outside world. At the gates of the pore protein, the chaperone hands over the subunit to enable it to connect to the last subunit in the growing hair.
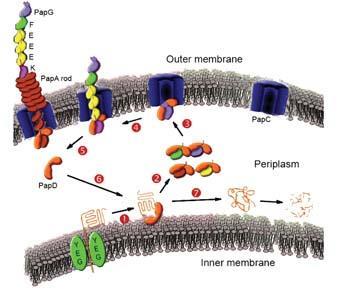
Emanuele Paci, a senior research fellow in the department of physics at the University of Leeds, in collaboration with Waksman's team, has produced molecular simulations which show that the 'zip-in-zip-out' mechanism (see Fig 2) is a plausible mechanism of the handover. This would imply that the β-sheet 'zipper' between chaperone and subunit gradually opens from one end, allowing the new zipper between the neighbouring subunits to close up and replace it.2
This highly complex pilus assembly machinery is specific to Gram-negative bacteria (which include many pathogens) and has no equivalent in higher organisms, and is therefore a potential target for new antibacterial therapies. Researchers are therefore looking for chemicals that will suppress the growth of these hairs and help fight infection, and at the same time offer a way out of the current crisis caused by drug-resistant bacteria, which seem to be spreading ever faster.
A promising strategy
There is a fundamental strategic difference between conventional antibiotics and antibacterial agents that suppress a virulence factor such as pili. Antibiotics kill sensitive bacteria fast and efficiently, which is good for the individual patient being treated, but may be bad for future patients, because it encourages bacteria to develop resistance genes to survive. If only one bacterium in a billion carries a plasmid (a small, ring-shaped piece of DNA) with the gene that makes it resistant against a given antibiotic, the widespread use of this antibiotic will aid the propagation of this variant and it may soon spread throughout the (surviving) bacterial population. This is why 'superbugs' like MRSA tend to arise in hospitals, where patients, bacteria, and antibiotics can all be found in a small space, and why drug companies need to develop new antibiotics all the time. It also explains why existing antibiotics lose their efficacy, especially if they are used carelessly.
By contrast, drugs targeting a non-vital virulence factor, such as the assembly of pili, will not kill the bacteria. While this looks like a weakness, it has two significant advantages. First, there is less chance that the bacteria will develop resistance genes, and even if they do it will only be at the site where the pili would normally anchor the bacteria, for example the urinary tract. For the survival of bacteria outside this location, the pili aren't necessary, so any drugs leaked into the environment don't breed resistance. Secondly, the 'disarmed' bacteria may remain present in the body for long enough to allow our immune system to learn how to fight them, so that they act as an immunisation against the disease.
Chemists to the rescue
With these potential advantages in mind, chemists have set out to develop pilicides, ie small molecules that can stop bacteria from growing pili. Previous efforts to block the adhesins from binding their targets were hampered by the diversity of the binders. Researchers were unable to find a universal blocker, and so turned their attention to the production of pili.
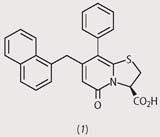
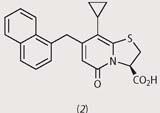
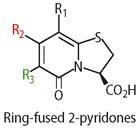
Fredrik Almqvist and his research team at the University of Umea, Sweden, have recently designed a new class of molecules which act as peptide mimics, with the hope that these could interfere with the zipping mechanism that connects subunits to each other. The Swedish researchers developed synthetic procedures (see Box)3 to produce a wide range of 2-pyridones with various substituents in the positions R1, R2, and R3.
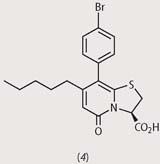
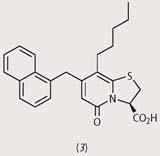
From a first screening, they identified four disubstituted pyridones (structures (1)-(4)), which had strong activity against pilus assembly. Using these four pilicides as lead compounds, the researchers went on to develop substances with improved characteristics such as solubility and uptake by bacteria.
In biological and biophysical tests, bacteria treated with some of these new pilicides showed that these substances interfere both with the chaperone and with the usher proteins, thus blocking the growth of the bacterial hairs and reducing their ability to cling to host cells. Clinical trials now need to be done to determine whether this effect can be extended to bacteria which cause infections in humans.
Rudi Glockshuber and his group at the ETH Zurich have successfully reconstituted the pilus assembly machinery in a test tube.4 Their system includes the adhesin, the bulk subunit, chaperone and usher proteins, and produces pili of comparable length as bacteria, and with the same speed, provided that the usher protein is first primed with the chaperone/adhesin complex, before being exposed to the chaperone/subunit pairs. Remarkably, the whole system turned out to work independently of cellular energy supplies. Such user-friendly model systems could help the early drug screening for new types of pilicides.
Michael Gross is a science writer based at Oxford and an honorary research fellow at the School of Crystallography, Birkbeck College, London. He can be contacted via his web page.
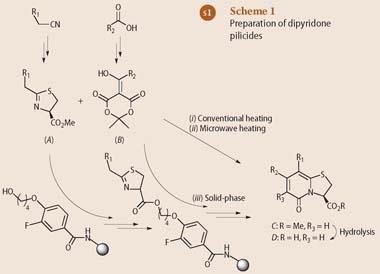
How to make dipyridones
Almqvist and coworkers prepared their dipyridone pilicides from two classes of building blocks, A, and B (see Scheme 1). The thiazolines, A, with various side chains R1 can be obtained from nitriles, including the corresponding functionalities. Similarly, compounds B with various side chains R2 are derivatives of Meldrum's acid (2,2-dimethyl-1,3-dioxane-4,6-dione) and can be produced by condensation of simple compounds such as carboxylic acids and ketones.
The researchers have described three different methods of combining compounds of the A and B type to form a dipyridone, C, namely by conventional heating, with microwave assistance, or in the solid phase.3 By hydrolysing the ester group, they obtained carboxylic acids D, which can serve as mimics of the carboxy terminus of a polypeptide chain.
Based on the structural framework of D, the chemists carried out further modifications, including halogenations, formylations, and nitrations to explore the effects of these additional functionalities. They also added methylamine groups to improve the solubility of the compounds obtained and tried attaching additional amino acid residues to mimic longer peptides.
They produced dozens of new compounds with a range of properties and activities. While most compounds were able to bind to Pap proteins, the researchers found that only those classes which (like D) have a carboxylic acid group mimicking the carboxy terminus will inhibit pilus growth in vivo.
Related Links
Michael Gross Science Writer
References
D. Verger et al, PloS Pathog., 2007, 3 (5): e73, doi:10.1371/journal.ppat.0030073.
R. J. Rose et al, J. Mol. Biol., 2008, 375, 908.
V. Åberg & F. Almqvist, Org. Biomol. Chem., 2007, 5 (12), 1827.
M. Nishiyama et al, Science, 2008, 320, 376.






No comments yet