Advances in mass spectrometry (MS) technology over the past 30 years have pushed this technique into the hands of biologists and biochemists, and led to a host of new applications.
-
The development of soft ionisation techniques has meant MS can be used to analyse large biological molecules
-
Protein analysis, the quality control of biofuels, and forensics all benefit from advances in MS technology
Mass spectrometry (MS) relies, among other things, on two fundamental criteria. The atom or compound of interest must be ionised to generate charged species, and these are then mass analysed to determine their mass-to-charge (m/z) ratio. From the m/z values, physicists have investigated the properties of stable isotopes and chemists have been able to deduce the structure of small organic molecules. Further, MS is increasingly finding application in solving biological problems.

MS - the basic principles
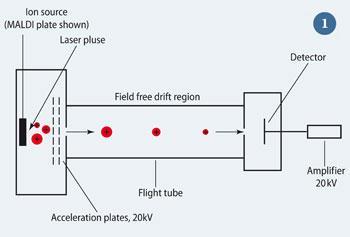
In a mass spectrometer, the sampleto be analysed is ionised by, for example, bombarding it with an electron beam, ie electron ionisation (EI). The ions formed are then accelerated by an electric field and pass through a mass analyser.
The time-of-flight (TOF) mass analyser (see Fig 1) uses an electric field to accelerate the ions to the same kinetic energy ( = 0.5mv2), and measures the time they take to reach a detector at the end of the tube. The following relationship applies:
t = k (m/ z)1/2
where k is a constant. Thus the lower the m/z of an ion, the faster it will reach the detector. As the initial kinetic energy and time are known, the m/z, and thus the mass of the ion can be determined.
Making macromolecules fly
The 1980s saw the development of three 'soft' ionisation techniques, which brought MS into the hands of biologists and biochemists. These techniques - fast atom bombardment (FAB); matrix-assisted laser desorption/ionisation (MALDI); and electrospray ionisation (ESI) - impart little energy to the molecule during the ionisation process and thus cause little or no fragmentation. Further, the molecules do not have to be in the gas phase prior to ionisation. Thus thermally unstable molecules of biological relevance could now be analysed using MS.
The first of these techniques, FAB, was pioneered by Michael Barber and colleagues at UMIST in Manchester,1 and allowed molecules such as peptides and carbohydrates to be analysed without prior derivatisation. This approach was superseded by MALDI2 and ESI,3 which are routinely used today.
MALDI
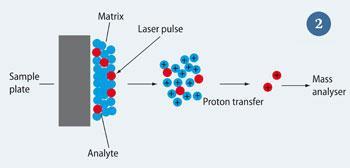
MALDI was developed by Michael Karas and Franz Hillenkamp at the University of M?nster, Germany (see Fig 2) as a variant of soft laser desorption, the Nobel prize-winning technology developed by Koichi Tanaka.4 The sample is first dissolved in a weak organic acid (the matrix), which is normally in excess of the analyte. The resulting solution is then spotted onto a sample plate and irradiated with a laser. Typically, a laser with a wavelength in the uv region of the electromagnetic spectrum is used. The matrix absorbs at or near the laser wavelength, which causes the matrix molecules to sublime from the surface. Analyte molecules are carried with them and ionisation takes place in the gas phase immediately above the sample plate. This is achieved by proton transfer, leading to predominantly
[M + H]+ species being produced. The ions are then sampled into the mass analyser. Using MALDI, Karas and Hillenkamp demonstrated that it was possible to ionise and detect bovine albumin, a protein with a mass of 66430 Daltons (Da).5
ESI
ESI was developed as a method of interfacing MS with liquid chromatography by John Fenn and coworkers at Yale University, US.3 The technique involves dissolving the sample in a polar solvent, typically water, methanol or ethanenitrile (acetonitrile). The solution is passed through a metal capillary to which a high voltage, usually between 3-6 kV for positive ESI, is applied (Fig 3). A counter electrode is held at 0 V, causing a strong electrical field to be created. Consequently, the solution comes out of the capillary as small droplets, which become smaller as solvent molecules evaporate, eventually leaving the isolated ions to enter the mass spectrometer. This sample molecule is either ionised by protonation, ie [M + H]+, or cationisation, eg [M + Na]+. Conversely, negative ESI can be performed where a proton is abstracted from an acidic group on the analyte by using negative voltages.
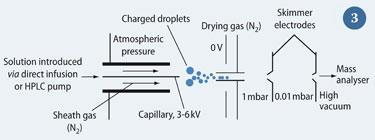
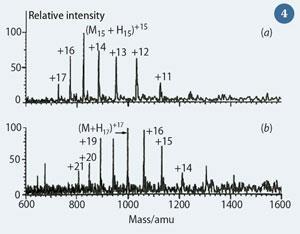
A benefit of ESI is that multiply-charged species are observed ie [M + n H]n+ Macromolecules, such as peptides and proteins, often have a large number of basic sites capable of accepting a proton. This leads to a range of species existing in solution, differentiated by the number of protons attached to the molecule. When the solution is electrosprayed into the mass spectrometer, these species are recorded at different m/z values because the m value stays almost constant while z varies. The resultant distribution of peaks is referred to as the 'charge envelope', see Fig 4.6 The reduction of the m/z value of a molecule through multiple charging means that macromolecules can be analysed on mass spectrometers that have a low upper m/z range limit, such as quadrupoles, increasing the utility of such instruments.
The annotations (Fig 4) show the number of protons associated with each peak. Deconvolution of the spectrum allows the molecular weight of the protein analysed to be determined.
Fenn was awarded the Nobel Prize for chemistry in 2002 in recognition of the immense impact that ESI had on the scientific world. He demonstrated that large molecules in excess of 130,000 Da could be ionised using his new technique.6 The combination of MALDI and ESI paved the way for structural biologists to analyse large proteins to understand how biological systems work.
DESI
A variant of ESI - desorption electrospray ionisation (DESI) - was developed in the laboratory of Graham Cooks at Purdue University, US.7 This was the first of a series of 'ambient' ionisation techniques, which ionise samples outside of the mass spectrometer. This makes it easier to manipulate the sample during analysis and makes in situ MS a real possibility in the future. The analyte need not be dissolved in a matrix (as in MALDI), thus negating the need for sample preparation.

In DESI, the same solvents as used for conventional ESI are electrosprayed onto the surface containing the analyte of interest. Upon contact with the surface, the droplets transfer energy, causing the molecules to desorb and become ionised. The same types of ions as conventional ESI, including multiply-charged species, can be observed.
An advantage of DESI is that the removal of the sample preparation step makes the technique suitable for high-throughput analyses, such as checking luggage for traces of drugs and explosives at airports.8
The advances in MS have not just been restricted to ionisation techniques. Mass analyser technology has developed, leading to faster and more selective instruments. As such analysts can detect more compounds and have greater confidence that they are identifying them correctly. Further, mass spectrometers can now
have limits of detection in the low attomole (10-18 moles) region.9
Sorting the compounds

An important analytical approach often coupled to MS is chromatography.10 Chromatographic methods can allow the separation of complex mixtures so that each component enters the mass spectrometer at a different time. This ensures that the spectra recorded are not composites of more than one compound. This is especially important for modern mass spectrometric analyses where complex matrices of potentially thousands of compounds are encountered. Bodily fluids in biological MS and mixtures of hydrocarbons in biofuel analysis are two examples.
The most common chromatographic methods are gas chromatography (GC) and high-performance liquid chromatography (HPLC). The chemistry of the analyte of interest determines which approach is used, but both methods can easily be coupled to MS. The principles of separation in both techniques are similar with two 'phases' being employed. The instrument consists of a column to which a stationary phase is bonded. The gaseous or liquid mobile phase containing the sample then passes over it. The different compounds have different affinities for the two phases, so that those with a greater affinity for the mobile phase elute from the column into the mass spectrometer earlier than those with a greater attraction to the stationary phase. Thus, separation is achieved.
Applications

The development of MALDI and ESI has increased the use of MS in the biological sciences, and has fuelled the '-omics' revolution. Analogous to the Human Genome Project, where the genes of the human genome were mapped, '-omics sciences' aim to identify the full complement of a target type of compound in a biological system. Examples include metabolomics, where endogenous metabolites are profiled, glycomics, which involves identifying the entire complement of sugars, and proteomics, the analysis of proteins in biological systems.
A typical proteomics experiment involves analysing the proteins present in a diseased tissue (for example, the cells of a cancerous organ) and comparing the results with those from a healthy, control sample. The increased or decreased expression of certain proteins can be used as biomarkers for the disease in question.
Mass spectrometry is ideally suited to this task, owing to its low limit of detection. This is important because some proteins, for example, are present at femtomolar quantities (1 femtomole = 10-15 moles), equivalent to around 100 copies of the molecule per cell in the amount used in a typical proteomics experiment.
Mass spectrometry can also be used to fragment ions to deduce information about their structure using the tandem mass spectrometry (MS/MS) approach.9 Several MS/MS experiments exist but the common theme is that fragmentation of the protonated molecules is induced. The sequence information in a proteomics experiment can be derived from either the protein itself or its constituent peptides. 'Top-down' proteomics requires fragmentation of the protonated protein, whereas a 'bottom-up' experiment involves digesting the protein with an enzyme, typically trypsin, and then performing MS/MS on the resultant protonated peptides.
Either approach will produce fragment ions that are related to the sequence of amino acids constituting the peptide or protein. The sequence can then be deduced from first principles or by reference to a database to identify the protein.
MS for a green future

The most common biofuel in Europe is biodiesel, which is most frequently blended with petrodiesel. 11 The ability of MS to deliver both qualitative and quantitative data in a single experiment makes it ideal for the quality control of biodiesel.
The mass spectrometer is interfaced to a gas chromatograph, essential owing to the complex nature of petrochemical matrices, and can be used to monitor both the FAMEs (fatty acid methyl esters, the constituents of biodiesel) in the sample as well as any impurities and degradation products that could damage or clog the engine.
The older technology of EI is particularly useful, producing diagnostic fragmentation patterns that can be compared against databases to identify the compounds in the biodiesel. By using the intensities of the signals produced by the compounds in the sample and comparing them to that generated for an internal standard, an accurate estimate of each compound can be made using GC-MS. This is particularly important because UK legislation requires all diesel to be 5 per cent biodiesel by 2010.
Seeing pictures with MS
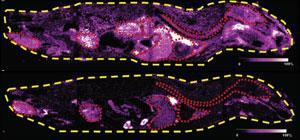
The concept of 'imaging MS' was introduced by Richard Caprioli in 1997.12 The approach allows an image of a sample to be produced, which shows the distribution of compound(s) of interest. A common imaging MS experiment involves the detection of a biologically relevant compound in a tissue. In an example, thin slices of rat brain were made after the drug olanzapine had been administered.13 MALDI-MS/MS was performed to acquire product ion mass spectra of a diagnostic fragment ion of olanzapine (Fig 5). The images were obtained by taking spectra across the whole tissue section, thus producing a mass spectrum at each x, y coordinate. By converting the ion intensity to colour at each point, the distribution of olanzapine was shown to have decreased in the brain and spinal cord after six hours compared to after two hours. This showed that the drug was cleared from the central nervous system in this time period.
Imaging MS can also be performed using DESI.14 The technique has been used to produce images of latent fingerprints (Fig 6). These not only give pattern information comparable to a traditionally imaged fingerprint eg with powder, but also provide evidence about which chemicals the person has been in contact with. The image in Fig 6 was produced by acquiring mass spectra of the protonated molecule of cocaine after just 5μg of the compound had been smeared across the subject's finger.

Conclusion
The applications of MS here are merely the beginning of the story. MS is increasingly finding application in the understanding of pharmaceutical drug metabolism, in toxicological screening for drugs of abuse in athletes, in the measurement of heavy metals in environmental samples and many other areas. It is anticipated that increased usage of existing MS technology, coupled with new instrumental developments, will see the use of the technique become even more common in the future.
Stephen W. Holman is post-doctoral research associate in the Michael Barber Centre for Mass Spectrometry at the University of Manchester, 131 Princess Street, Manchester M1 7ND.
Further Reading
For definitions of terminology used in this article, visit the IUPAC mass spectrometry terms project weblink
References
1. M. Barber et al, J. Chem. Soc. Chem. Comm., 1981, 325.
2. M. Karas et al, Int. J. Mass Spectrom. Ion Processes, 1987, 78, 53.
3. C. M. Whitehouse et al, Anal. Chem., 1985, 57, 675.
4. K. Tanaka et al, Rapid Commun. Mass Spectrum, 1988, 2, 151.
5. M. Karas and F. Hillenkamp, Anal. Chem., 1988, 60, 2299.
6. J. B. Fenn et al, Science, 1989, 246, 64.
7. Z. Takáts et al, Science, 2004, 306, 471.
8. R. G. Cooks et al, Science, 2006, 311, 1566.
9. E. de Hoffmann and V. Stroobant, Mass spectrometry: principles and applications (3rd edn). Chichester: John Wiley and Sons, 2007.
10. C. Poole, The essence of chromatography. Amsterdam: Elsevier, 2003.
11. G. Knothe, J. Van Gerpen and J. Krahl (eds), The biodiesel handbook. Urbana: AOCS, 2005.
12. R. M. Caprioli, T. B. Farmer and J. Gile, Anal. Chem., 1997, 69, 4751.
13. S. Khatib-Shahidi et al, Anal. Chem., 2006, 78, 6448.
14. D. R. Ifa et al, Science, 2008, 321, 805.









No comments yet