German researchers develop a simple approach to making rigid DNA nanorings with tailor-made functionality
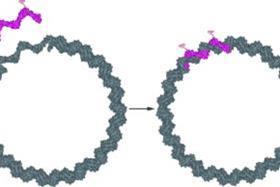
A simple approach to making rigid DNA nanorings with tailor-made functionality has been developed by Michael Famulok and his team at the University of Bonn, Germany.1 The development could open up new pathways for the construction of highly organised, DNA-based objects for preparing nanocomposite materials.
Famulok's nanorings reveal themselves under the atomic force microscope to be composed of double-stranded DNA with a tiny gap in the form of a short single-stranded fragment. Altering the nature of the single-strand fragment offers a way to design nanorings with specific functionality.
Nucleic acids are fascinating building blocks for nanocomposite materials because they are easy to make with various molecular recognition characteristics. The difficulty in using these materials lies in creating defined two- or three-dimensional geometries, which usually requires rigid building blocks, whereas DNA molecules are generally flexible structures.
'From the structural point of view, miniature rings represent the simplest form for a rigid object made of DNA', explains Famulok. His team has found a way to produce DNA molecules with a smooth circular structure, free of ring deformations, twists, or knots, by carefully selecting the sequence and number of nucleotides as well as by exploiting a neat synthetic route that leads to the desired rigid rings.
References
- M. Famulok et al, Angew. Chem. Int. Edn, 2008, 47 (5), 967.






No comments yet