If computer technology develops at the rate it is developing today, in just 10 years computers would be expected to be 1000 times more powerful, and hard disks 10,000 times more spacious. But the miniaturisation of silicon chips is fast reaching its limit.
- Chemists copy Nature in attempt to build computers atom by atom, molecule by molecule
- Molecular self-assembly will be key to engineering nanoscale computers

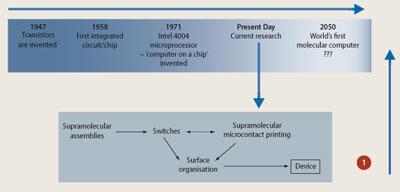
What we have achieved to date in computer technology has been through a 'top-down' approach where materials and devices are built up by removing existing material from larger entities.1 Lithography, for example, is used in the production of silicon-based chips, but such technology cannot down-size electronic components to atomic magnitudes that will be required for tomorrow's faster and more powerful computers.2 For this researchers are turning to a 'bottom-up' approach that relies on the self-assembly of molecular structures. This will involve building molecular-scale components atom by atom, which3 will allow chemists to exert control over the dimensions and fundamental properties as well as composition of computing components (see Fig 1).4 The idea, perhaps not surprisingly, comes from Nature.
Lessons from Nature
In Nature when chemical building blocks of the right type come together in different combinations we see structures of various size and complexity. Proteins and viruses are examples, as is DNA. The self-assembly of DNA is a 'bottom-up' process that involves the coming together of particular types of building blocks, ie nucleic acids. It is the arrangement of these building blocks that gives rise to the uniqueness of each strand of DNA. Nature, by carefully controlling the properties of the various building blocks, is able to facilitate the self-assembly as well as step-wise growth of complex systems and thus bypass problems associated with pure step-wise construction. The result is that, by holding the building blocks in the right way through weak interactions and covalent bonds, Nature can assemble myriad complex structures.
The key to understanding self-assembly and thus realising the ultimate goal of assembling nanoscale computers by using a 'bottom-up' strategy will lie in the design of the building blocks and how these can be held together to build larger architectures. Spontaneous self-assembly of molecules or 'supramolecular chemistry' will depend on:
- thermodynamic (strength of the interactions) and kinetic factors (speed of assembly);
- the type of building blocks used;
- the forces that hold the structure together, both weak and reversible.
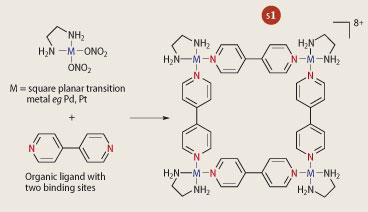
The interplay between molecular forces eg hydrogen bonding, π-π stacking and electrostatic interactions should be able to generate structures of amazing proportions and diversity, and it is in these forces that the power of supramolecular chemistry will lie. In theory if we were to place all the different building blocks, organic and inorganic, into 'one-pot' we should be able to generate a variety of molecular architectures, which could go on to form one dimensional chains, two dimensional networks and three dimensional arrays. Thus, the building block concept will be vital to producing a 'nano-device' that will build itself. By using weak interactions, the building blocks will have time to 'error-correct' to the right architecture if the wrong one is formed. By carefully selecting organic ligands with multidentate binding sites we should be able to assemble structures that can grow in three dimensions or can be limited to growing in only one direction.
If we take, for example, a transition metal with square planar geometry, and add four ligands (also known as 'spacers') that contain two binding sites each, we can assemble a molecular square (see Scheme 1). By replacing the 'spacer' unit with longer 'spacers' which contain more binding sites, we should be able to produce highly complex structures. In fact, the library of complexes that exist based on this type of assembly is increasing and has allowed researchers insight into understanding molecular organisation. They can already exert a degree of control over producing not just novel structures, but structures that contain the required functionality, which could be used in molecular electronics for the future's computers.
Building molecular computers
Before we can exploit this technology in computing, however, we need to understand how a computer works. Modern computers have three key parts - input devices, output devices and memory. The component common to each of these and the one researchers are most interested in is the transistor, the smallest physical part of a computer processor. The key to the next generation of computer technology will lie in our ability to miniaturise the transistor. (It's worth remembering that the first valves measured ca 14 cm (height) x 5 cm (diameter) and today's transistor is 40 μm across.) The transistor itself has three regions - a base, a collector and an emitter. When current flows between the collector and emitter the transistor is 'on', representing that the voltage to the base has increased above a threshold. When the voltage at the base is less than the threshold the transistor is in the 'off' state. The transistor is a switch because it has the ability to change from the 'on' and 'off' state.
Based on the same principles, but scaled down to the molecular size, scientists are currently developing molecular devices that can perform switching functions. In theory, a molecule that can change between two distinctive states can be used as a molecular switch and thus be used to build transistors for molecular computers and storage devices. Rotaxanes, for example, are supramolecular assemblies made of several different components. Arranged onto a linear unit (the 'molecular thread') is a macrocyclic ring with two bulky substituents (the 'stoppers') at either end (see Fig 2 (a)). As a result of this, these molecules are often described as having a 'dumb-bell' shape.5 The linear unit is made of an organic polymer, which can be functionalised at both ends with bulky organic groups to act as the 'stoppers'. The macrocyclic ring that encircles the 'thread' can be held in position by weak interactions such as hydrogen bonding, hydrophobic forces and coordination bonds.
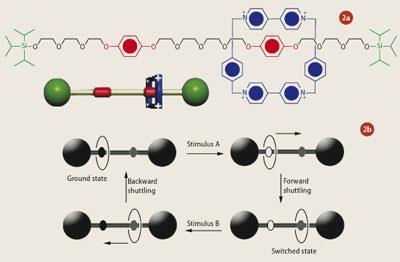
When external stimuli such as photochemical, electrochemical or chemical energy are applied to the molecule, the 'ring' shuttles along the 'molecular thread' stopping at either one of two recognition sites located along the thread (see Fig 2 (b)).6 Thus the response of rotaxanes to such external stimuli gives rise to two distinct physiochemical states, which makes rotaxanes ideal contenders for use as molecular switches.
Other molecules that have the potential to be used as molecular switches are metal oxide clusters.7 These clusters represent ideal molecular components, owing to their vast structural diversity and wide range of physical properties such as magnetism, conductivity and photochemical activity. They have the added advantage of being able to encapsulate or protect the computing component inside the cluster molecules.
Recently a molybdenum-based cluster has been designed that changes colour when stimulated.8 Encased inside the molybdenum-oxygen metal cage are two sulfite anions arranged in a precise environment. Activation results in the cluster changing its electronic state and thus its colour. The cluster unit has the potential to act as a storage device or as a single molecule transistor.
Self-assembling arrays
Although it is possible to build precise molecular systems that can formally switch, to be of any use in molecular computers the components need to be connected or arranged in an environment so that they can be located and addressed. One answer to this problem could lie in the construction of connected grids. Two dimensional arrays based on grid-like assemblies have recently been the focus of much research (see Fig 3).9 These grid-like architectures have many key features - eg redox, magnetic, and spin-state transitions - that make them ideal candidates to form integrated circuits. Furthermore, they resemble the cross bar architectures present in current integrated circuits, which are used to store and process data.9 Finally, a number of these architectures can be deposited onto solid-state surfaces through modern scanning probe techniques such as STM (scanning tunnelling microscopy - using conducting electrons to probe) and AFM (atomic force microscopy using force measurement to probe). The recent advances in these techniques have allowed researchers to not only deposit molecular materials onto solid-state surfaces but also to manipulate them on the surface.
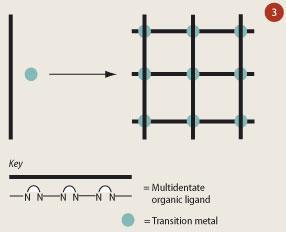
Ultimately, all of these molecules represent the building blocks that have to be integrated into circuits to fulfil the fantasy of nanoscale computing devices. Supramolecular chemistry can then assemble these building blocks to form the nanostructures that researchers aim to make. What would be ideal is if molecules that are switches could by self-assembly position themselves onto a surface at the right location.10
Putting it all together
We are now reaching the end of our conceptual journey and so far we have shown how molecular assemblies can have designed function and be used like transistors, and how these assemblies can build themselves as long as the chemist designs the building blocks correctly. However, the major problem remains in understanding how to fabricate arrays of molecular architectures that can communicate with each other and with the macroworld.
Essentially what is required is 'focused self-assembly' where all the molecular components can be directed into the right location and orientation to generate integrated nanosystems. To do this we will need to utilise existing engineering approaches with self-assembly and surface chemistry.
Electrical engineers and physicists make nanostructures using lithography, whereby patterned structures are created with dimensions as small as 45 nm. The processors that are present in our computers today are made in this way. There are various types of lithography but in general the process involves the output of data that creates a pattern on the substrate surface.11 Although a 'top-down' approach, the advantage of lithography is that it can make regular arrays of nanostructures, something which is not yet conceivable with self-assembly. However, it is possible that surfaces patterned with organic molecules may be used as templates for the assembly of molecules.
Self-assembled monolayers (SAMs)12 represent a technique that can realistically combine the current lithographic techniques with self-assembly. Several research groups have developed the concept of a 'molecular printboard', which is made from a monolayer of molecules deposited onto a solid surface. These molecules act as the 'hosts' to which 'guest' molecules can attach through supramolecular interactions, in a controlled way. For instance, researchers have recently designed SAMs that incorporate molecular receptors, which can bind other molecules at the surface selectively.13 In this system the receptors, ie the 'hosts', can recognise the 'guests' because they contain molecular cavities, thus enabling the controlled positioning of the 'guest' molecules. Techniques such as supramolecular microcontact printing and supramolecular dip-pen lithography are used to arrange molecules onto the 'molecular printboard'. The most commonly studied systems use alkanethiolates, which can spontaneously chemisorb onto a gold surface - there is a strong affinity between the sulfur groups and the gold surface. The molecules that are placed onto the gold surface also contain terminal groups which can be modified to give the required terminal functionality to interact with other molecules that may be deposited onto the surface.
What scientists now need to do is to use such techniques to direct the assembly of molecular switches at specific positions, thus not only achieving the self-assembly of molecular arrays but in turn generating integrated systems. Using various lithographic techniques, substrates can be patterned with molecules at regular intervals that will almost act as 'homing beacons', directing or encouraging the molecular switches to self-assemble or 'anchor' at these points.
And finally
The key to realising molecular computers lies in our understanding of how Nature assembles functional structures with such complexity and purpose. The ability to compress more computing components into a given area is a challenge for the nanoscale chemist. However, the biggest challenge will be to solve the problem of combining 'top-down' and 'bottom-up' methodologies. This will have revolutionary implications as to what will be achievable at the nano-macro world interface, and in turn will open the door to designing more powerful computational devices.
Lee Cronin is professor of chemistry in the department of chemistry at the University of Glasgow, Joseph Black Building, University Avenue, Glasgow G12 8QQ; Hamera Abbas is a PhD student in the same department.
References
- N. Robertson and C. A. McGowan, Chem. Soc. Rev., 2003, 32, 96.
- D. Appell, Nature (London), 2002, 419, 533.
- D. Cobden, Nature (London), 2001, 409, 32.
- L. Samuelson, Mat. Today, 2003, 22.
- (a) P. L. Anelli et al, J. Am. Chem. Soc., 1992,114, 193; (b) F. M. Raymo and J. F. Stoddart, Chem. Rev., 1999, 99, 1643.
- H. Tian and Q.-C. Wang, Chem. Soc. Rev., 2006, 35, 361.
- D.-L. Long and L. Cronin, Chem. E. J., 2006, 12, 3698.
- D.-L. Long, P. Kögerler and L. Cronin, Angew. Chem. Int. Edn, 2004, 43, 1817.
- M. Ruben et al, Angew. Chem. Int. Edn, 2004, 43 , 3644.
- R. F. Service, Science, 2002, 295, 2398.
- M. Geissler and Y. Xia, Adv. Mater., 2004, 16 , 1249.
- B. D. Gates, Chem. Rev., 2005, 105 (4), 1103.
- O. Crespo-Biel et al, Dalton Trans., in press.






No comments yet