Microorganisms have evolved the ability to synthesise molecules - siderophores - that allow them to acquire essential iron from their environment. Owing to their remarkable affinity and selectivity for iron(III), siderophores have recently attracted renewed interest, mainly from medicinal and analytical chemists who are looking to use them as drugs and as sensors.1
- Siderophores contain oxygen atoms, which bind to Fe(III), making it water soluble and allowing cellar uptake
- Antibiotics attached to siderophores could provide the solution to microbial resistance to these drugs
The atmosphere that supported the Earth's first living organisms was very different to the one we live in now. In contrast to the oxygen-rich atmosphere of today, billions of years ago the Earth's atmosphere was reducing in character, containing ammonia, carbon dioxide, methane and hydrogen sulfide in addition to nitrogen. Metal ions were mainly present in their lower oxidation states, typically existing in the form of sulfides.
Under these conditions, iron(II) was soluble and readily available and so became incorporated into organisms as a biometal, which could catalyse redox processes in the active sites of many proteins and enzymes.
The evolution of siderophores
As oxygen began to accumulate in the atmosphere, owing to photosynthesis, the availability of iron changed drastically. In the presence of oxygen, reasonably water soluble iron(II) sulfide was oxidised to iron(III) oxide and hydroxide, becoming insoluble at neutral and basic pH.
As a consequence, microorganisms had to find a new way of obtaining iron from their environment. They evolved molecules known as siderophores - from the Greek word for 'iron carriers' - which bind to iron(III), making it water soluble and thereby enabling cellular uptake.
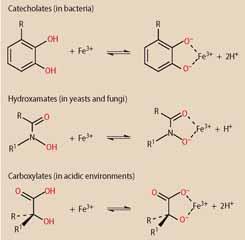
To bind to iron(III), siderophores rely on oxygen donor atoms because these have a high-affinity for small, highly-charged metal cations. According to the type of oxygen donors used, siderophores can be divided into different classes:
- catecholates;
- hydroxamates; or
- carboxylates.
In general, bacteria produce catecholate siderophores while fungi tend to rely on hydroxamate siderophores. Since carboxylate siderophores bind to iron(III) even at low pH, they are produced by microorganisms living in acidic environments.
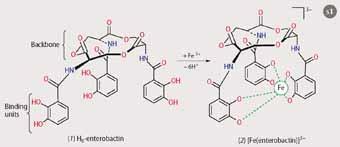
Iron(III) has a preference for being six-coordinate, so the most efficient siderophores have three didentate binding units. Enterobactin of Escherichia coli, for example, has three catecholate units attached to a cyclic backbone, which holds the units in the right position to 'encapsulate' the iron. Upon iron(III)-binding, the protons on each of
the oxygen donor groups are released, resulting in the highly stable and water soluble 1:1 complex, [Fe(enterobactin)]3- (see Scheme 1).2
The iron(III)-siderophore complex is recognised by specific receptor proteins in the outer membrane of the bacterial cell and actively transported inside. A receptor protein will only recognise its respective complex, ie some only transport iron-complexes of catecholate-type siderophores. The receptors recognise the shape and electronic character of the coordinated binding units, initiating transport into the cell, similar to a key unlocking a gate.
Siderophores in medicine
Over time, many antibiotics have become less effective in treating bacterial infections because the bacteria have mutated and developed resistance. Consequently, chemists are looking for novel ways of combating bacterial infections.
One approach is to use a 'Trojan Horse' strategy.3 This involves attaching the antimicrobial agent to a siderophore, so that the antibiotic is taken up along with the siderophore when the bacterium is sequestering iron. The backbone of the siderophore is not directly involved in the recognition of the iron complex by the receptor protein, so it can be attached to an antibiotic without affecting cellular uptake. In this way, the antimicrobial agent can be smuggled into the cell. If the bacteria mutate to block the entry of the siderophore-linked antibiotic, they will suffer from iron starvation and die. There is no reason why even highly resistant bacterial strains or 'superbugs' would not respond to such a strategy.
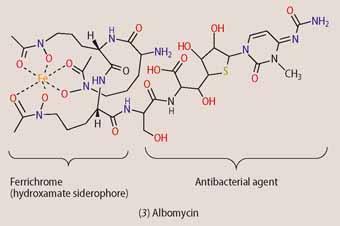
Some microorganisms produce such siderophore-linked antibiotics to gain an advantage over competing bacteria. Albomycin (3) is an example of a natural 'Trojan Horse', which is produced by Streptomyces griseus. Albomycin consists of the hydroxamate siderophore ferrichrome, which is connected to an antibacterial agent via an amide linker. Now, though E. coli does not produce ferrichrome, it has developed receptors for it so that it can take advantage of it. To prevent E. coli from using its siderophore, S. griseus has evolved so that it can attach a molecule that is toxic to bacteria onto ferrichrome. Once inside the E. coli cell, the amide-linker between the ferrichrome and the toxic part is cleaved enzymatically and the active antibacterial agent is released.
Inspiration from the structures of natural siderophore-antibiotic conjugates, such as albomycin, has led to the synthesis of a range of siderophore-linked drugs, many of which have shown a higher antimicrobial activity than the respective antibiotic on its own.
Unexpectedly, however, the development of drug resistance remains a concern, because most bacteria can use more that one type of iron uptake pathway. If E. coli, for example, mutates and shuts down the hydroxamate siderophore transport system, the uptake of albomycin is prevented, but the cells can still take-up iron by using catecholate or carboxylate siderophores.
To overcome this problem, synthetic siderophores with mixed binding sites, eg catechol-hydroxamate hybrids, are now being investigated, because mutations to block both iron-uptake systems would make the bacterial cells much more susceptible to iron starvation. None of these synthetic 'Trojan Horse' antibiotics, however, has yet reached drug status.
Treatment of iron overload
Despite being an essential element for humans, we are unable to absorb iron efficiently from food or excrete it at a rate greater than one milligram per day. Usually, these two factors balance out, but some medical conditions result in the accumulation of iron in the body, especially in the liver, heart and pancreas. Excess free iron is toxic since it leads to the production of highly reactive hydroxyl radicals (see Scheme 2)4 which can damage these vital organs. Unfortunately, the early symptoms of iron overload, such as abdominal discomfort, fatigue and weight loss are relatively non-specific and therefore iron overload is often not recognised until substantial damage has occurred.
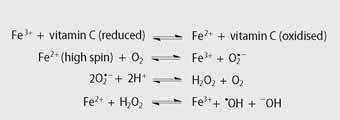
Chronic iron overload is a common side effect in patients who suffer from β-thalassaemia, a genetic blood disorder that leads to the production of a less effective form of haemoglobin. These patients require regular blood transfusions. Since each unit of blood (ca 0.45 l) contains approximately 250 milligrams of iron, regular transfusions lead to iron overload and ultimately iron poisoning. 5
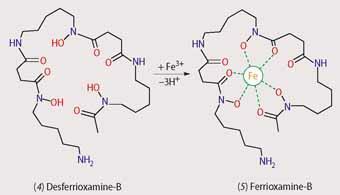
Currently, the most commonly prescribed drug for treating iron overload is the hydroxamate siderophore desferrioxamine-B (4), Scheme 3, marketed as Desferal, which was first isolated from Streptomyces pilosus. While this treatment is effective, there are several significant drawbacks - it is expensive, it is ineffective when taken orally and it rapidly degrades in the bloodstream. Consequently Desferal has to be delivered through a needle under the skin for a period of nine to 12 hours several times a week and, understandably, patient compliance is low. As a result, research has focused on developing orally-active siderophore mimics.
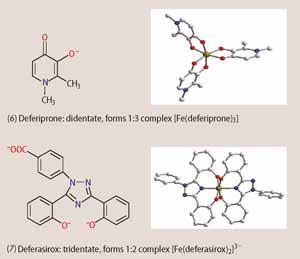
Recently, two orally-active drugs for treating iron overload have reached the market - deferiprone (6, Ferriprox) and deferasirox (7, Exjade). Deferiprone mimics the didentate binding units found in siderophores. It is isoelectronic to catecholates, but deprotonates only once upon iron binding, resembling the hydroxamates in this respect. Since deferiprone acts as a didentate ligand, three drug molecules are required to encapsulate the iron(III) in the form of a 3:1 complex. The three didentate ligands are not connected to a backbone, as in enterobactin or desferrioxamine-B, so they can dissociate more easily from the metal and open up coordination sites on the iron(III) centre, leading to the generation of toxic hydroxyl radicals as shown in Scheme 2.
Concerns about toxic side effects caused by the limited stability of the complex have unfortunately compromised the general introduction of the drug. In Europe, however, deferiprone was approved in 1999, but only as an alternative treatment for patients who experience problems with desferrioxamine-B.
Deferasirox on the other hand contains a nitrogen donor in addition to two phenolic oxygen donors. The drug acts as a tridentate ligand and two molecules occupy all six co-ordination sites of the iron(III) centre. The resulting 2:1 complex has greater thermodynamic and kinetic stability than the iron(III)-deferiprone complex. The US Food and Drug Administration (FDA) approved deferasirox worldwide for treating chronic iron overload in 2005. The drug is administered as a drink once a day and could make a significant difference to the lives of patients with chronic iron overload.
Molecular sensors for iron(III)
Selective molecular sensors that can indicate the concentration of iron(III) in the presence of other metal ions have many potential applications, ranging from medical analysis and biochemical research6 to environmental monitoring. The most selective molecular sensors developed to date have been those that mimic siderophores to optimise the affinity and selectivity for iron(III).

The simplest analytical method relies on the characteristic wine-red colour of iron(III)-catecholate siderophore complexes,7 which can be detected and measured by using UV/VIS-spectroscopy. Unfortunately, this method lacks sensitivity since the wine-red colour of the complexes is not very intense. In addition, the presence of other similarly coloured compounds within the sample can give misleading results.
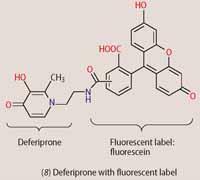
To detect low iron(III) concentrations and to minimise interferences, fluorescent labels have been attached to the backbones of siderophores or siderophore mimics, such as deferiprone. With fluorescent sensor molecules such as (8), the intensity or wavelength of the emitted light shows characteristic changes on iron(III) binding to the siderophore. Once bound to iron(III) and taken up by the bacteria, the fluorescence from the molecular sensors inside the cells can be detected by fluorescence microscopy, revealing the intracellular distribution of the iron-complex (8). By altering the type of siderophore, we can use these fluorescent sensors to visualise different species of bacteria. By taking advantage of microbial iron scavengers, we can adapt probes for various analytical or clinical purposes, with new applications expected to emerge alongside advances in imaging technology.
These relatively simple biomolecules have the potential to be versatile tools for synthetic chemists. They can be used directly as building blocks in the development of larger functional molecules or provide inspiration in the design of clinically or analytically useful iron scavengers.
Dr Anne-Kathrin Duhme-Klair is a senior lecturer in inorganic chemistry in the department of chemistry at the University of York, Heslington, York YO10 5DD; Emma L. Dux and Stephen J. Milner are PhD students in the same department.
References
- M. Miethke and M. A. Marahiel, Microbiol. Mol. Biol. Rev., 2007, 71, 413.
- S. K. Buchanan et al, Nat. Struct. Biol., 1999, 6, 56.
- J. M. Roosenberg et al, Curr. Med. Chem., 2000, 7, 159.
- W. Kaim, and B. Schwederski, Bioinorganic chemistry: inorganic elements in the chemistry of life. Chichester: John Wiley, 1994.
- P. V. Bernhardt, Dalton Trans., 2007, 3214.
- D. W. Domaille, E. L. Que and C. J. Chang, Nat. Chem. Biol., 2008, 4, 168.
- Y. Ma et al, Biochem. J., 2006, 395, 49.









No comments yet