In the near future, doctors will be able to carry out a 'while you wait' test, using genetic analysis, for chlamydia, the silent disease that can lead to infertility in women. This may be the start of a revolution in point-of-care medicine because detection of specific DNA sequences could, in principle, be applied to the diagnosis of just about any disease.

Genetic analysis, the detection of genomic nucleic acid sequences (see Box), is an important tool in modern science, having many applications. The detection of species-specific genes (short fragments of deoxyribonucleic acid, DNA) in host tissues can show the presence of pathogens or viral genes embedded in the host's genome. Infectious diseases such as HIV, Chlamydia trachomatis and hepatitis C can be diagnosed, as can bioterrorism agents in sterile body fluids.
In addition to the detection of DNA, RNA (ribonucleic acid) sequences can also be targeted. In the synthesis of a protein, a gene is copied as a single-stranded molecule (RNA). This messenger molecule, mRNA, moves to the part of the cell where proteins are synthesised (the ribosome) and acts as a template for the new protein. Detection of mRNAs can show which genes are being used, and at what levels. This can provide information about key processes that are occurring in an organism at a specific time. For example, increased expression of oestrogen receptor α is found in human breast carcinomas.
Mutations or changes to an individual's genome can lead to genetic diseases. Large genomic duplications and deletions (of the kilobase order) are the recognised cause of several inherited diseases, including β-thalassaemia (a serious form of anaemia), Duchenne and Becker muscular dystrophies (marked by weakness and wasting of selected muscles), and familial breast cancer. At the other end of the scale, slight mutations in an individual's genome (a change in just one base pair, for example) can lead to Huntingdon's disease, cystic fibrosis, and sickle-cell anaemia.
Sequence information from DNA
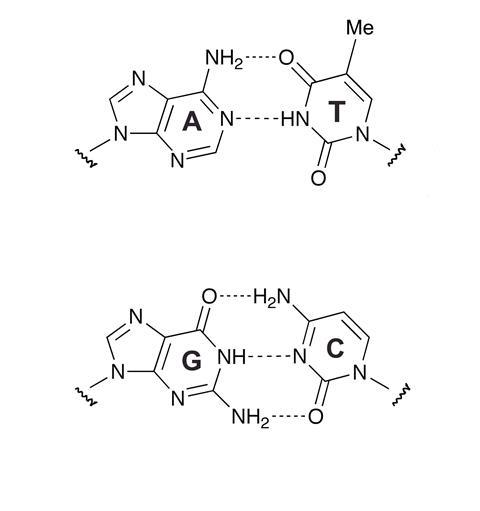
So how do scientists recognise a DNA sequence? The specificity of Watson-Crick base pairing controls the formation of duplexes between complementary sequences of nucleic acids (see Box). This fundamental process of molecular recognition makes it possible to target specific DNA or RNA sequences with oligonucleotide probes, ie short single strands of synthetic DNA or RNA strands of defined sequence. The formation of the AT (1) and GC (2) base pairs is a selective process, so to find out if any given nucleotide in the sequence is adenine (A), all we need to know is whether thymine (T) pairs with it. Similarly guanine (G) can be identified by its affinity for cytosine (C). Oligonucleotides are important tools in nucleic acid identification, amplification and isolation. If doctors need to know whether a patient has been infected with chlamydia, the presence of a pathogen-specific gene (DNA sequence) can be confirmed by determining if a complementary DNA sequence binds to it.
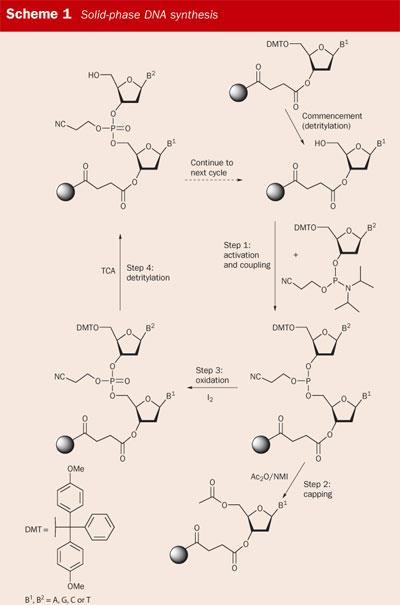
The ability to synthesise an oligonucleotide of any required sequence relies on solid-phase synthesis, a technique established by Bruce Merrifield for which he won the 1984 Nobel prize in chemistry. An oligomer such as a peptide or DNA strand is 'grown' on an insoluble solid support by using a series of chemical reactions. Throughout this process the oligomer remains chemically attached to the support and excess reagents are simply washed away. At the end of the assembly process the oligomer is cleaved from the solid phase and minimal purification is required to obtain a usable product.
Modern solid-phase DNA synthesis is based on pioneering developments by the organic chemists Marv Caruthers and Serge Beaucage at the University of Colorado in the early 1980s, see Scheme 1.1 Today, solid-phase DNA synthesis is so reliable that the yields for each reaction are typically >99 per cent. The whole process is automated: the chemist merely types in the sequence of bases required, loads the appropriate reagents onto the machine, and collects the product when the synthesiser has done its job. Perhaps a victim of its own success, the importance of DNA synthesis in genetic analysis is often overlooked as 'routine', precisely because of its efficiency.
Nucleic acid screening
Nucleic acid screening for diagnostic purposes requires amplification of a DNA sample followed by sequence recognition/identification by an appropriate oligonucleotide probe. Amplification is necessary to provide sufficient quantities of the sequence under investigation for detection because the original sample may contain just a few molecules of DNA.
Amplification
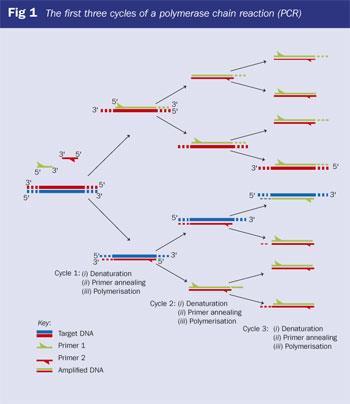
The polymerase chain reaction (PCR),2 the most powerful amplification technology available for producing large quantities of DNA from a sample, was developed by Kary Mullis. Driven to distraction by doing manual DNA synthesis at the Cetus corporation in the US (in the days before automation of Caruthers' chemistry), Mullis designed an incredibly simple way of making large quantities of DNA from a few initial copies. His method involves flanking the DNA region to be amplified (the DNA target or template sequence) with two synthetic oligonucleotide primers. Each primer is complementary to a region of one of the two different DNA template strands. The primers, typically 18-30 bases in length, are extended at their 3'-end during PCR by a thermostable DNA polymerase (typically Thermus aquaticus (Taq) polymerase) in a series of reaction cycles, each composed of three steps: denaturation, primer annealing and polymerisation (extension) (Fig 1). This process also requires the four different building blocks of DNA, the 2'-deoxynucleoside 5'-triphosphates or dNTPs - ie dATP, dGTP, dTTP and dCTP.
In the first cycle, the target DNA duplex is separated into two strands by heating to 95?C (denaturation). The temperature is then decreased to 55°C to allow the primers to anneal to the DNA target sequence. The temperature of the annealing step can be varied, depending on several factors, such as primer length and base sequence. After annealing, the temperature is raised to 72°C, the optimal temperature for polymerisation (extension step). During this step (requiring Mg2+), the template DNA is used by Taq polymerase to produce a complementary copy by extending the primers from their 3'-ends.
At the beginning of the second cycle, the PCR reaction is heated to 95°C again, and the process is repeated. The original target DNA and the newly synthesised strands both act as templates. In subsequent cycles, the newly synthesised DNA molecules soon outnumber the original target DNA molecules. In theory, one target molecule should generate 2n exact copies after n cycles, making PCR a very powerful amplification technique - 40 cycles generate 1,073,741,824 copies of a single template molecule.
A major bonus with PCR is that the amplified DNA is a specific region of DNA that lies between the binding sites of the two PCR primers. This means that if we start with the complete human genome (3 ×109 base pairs) only the region of interest is amplified (perhaps just 100 base pairs). This greatly simplifies subsequent DNA sequence analysis.
The specificity of PCR is attributed to the Watson-Crick base pairing during both hybridisation of the primers to the template and incorporation of the correct dNTP opposite the template base. The incorporation of the correct dNTP is controlled by the Taq polymerase. Taq polymerase is an enzyme isolated from an organism living in hot hydrothermal springs. Because of its origin, the enzyme is stable to the repeated heating encountered in the PCR cycle. Indeed, the development of thermostable polymerases based on Taq revolutionised PCR and converted it to a technique that can be used routinely in any lab.
Recently, with the emergence of real-time PCR, the importance of the technique has been reinforced in molecular genetics. In real-time PCR, amplification is monitored as the reaction occurs. This is in contrast to standard endpoint analysis and traditional methods such as sequencing, in which the presence and nature of the DNA sequence is determined by gel-electrophoresis after PCR or primer extension is complete.
Fluorescent labelling of oligonucleotides
To identify a specific PCR product we need a probe with a 'label' attached. Originally DNA probes were given radioactive labels; 32P was incorporated in internal or 5'-phosphate groups of DNA by enzymatic methods. Such probes, however, suffer inherent disadvantages: radioactive labels are hazardous, have short half-lives and short shelf-lives, are expensive, have generally limited signal emission, and the radioactive waste must be disposed of safely - all of which make them unsuitable for the GP's surgery.
In contrast, fluorescent molecules do not suffer from these disadvantages and have largely taken over as labels in nucleic acid technology. Fluorophores absorb light of a specific wavelength and re-emit it at a longer wavelength (lower energy). Incorporating these molecules into nucleic acids allows sensitive detection of the target DNA, provided that the signal is monitored at the correct emission wavelength of the fluorophore. Many fluorescent dyes are commercially available, with emission maxima covering the spectrum between ~400 and 800nm.3 The introduction of new hardware and detection systems has ensured that fluorescent detection can be done with a sufficient level of sensitivity for all relevant applications. Indeed, single-molecule fluorescence detection is currently a hot research topic.
Although the conjugation of fluorescent dyes to oligonucleotides is a relatively recent advance, the dyes that are used to label them have been in use for over a century. Fluorescein (3), by far the most common fluorescent label in real-time PCR, was first synthesised by Adolf von Baeyer, who was awarded the 1905 Nobel prize for chemistry in recognition of his work on organic dyestuffs. It will be familiar to many as the yellow-green fluorescent dye injected into the circulatory system to reveal retinal damage in a fluorescein angiogram, and its di- and tetra-brominated derivatives are used to provide the orange and deep red hues of lipsticks. Other widely-used labels are the cyanine dyes (Cy3, 4), which were used as sensitisers in the photographic industry as far back as the 19th century.
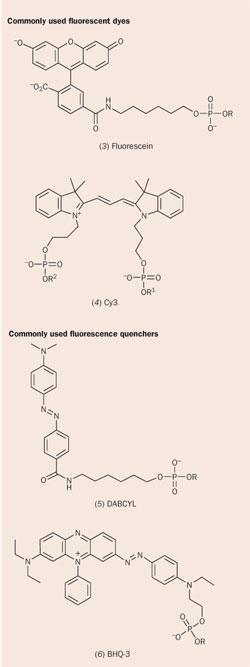
Real-time PCR with fluorescent oligonucleotide probes requires a 'signalling mechanism' that allows the level of fluorescence to indicate whether the probe has hybridised to its complementary sequence. Typically, this involves a change in separation between a fluorescent dye and a fluorescence quencher. Again, the quenchers are derived from 'industrial revolution' dyestuffs such as the azo dyes (DABCYL, 5) and rhodamines (TAMRA).
Although new, improved quenchers have been developed, the traditional ones are often the basis for their design - the 21st century 'Black Hole Quencher 3' (BHQ-3, 6) is an elaborated form of William Perkin's mauveine, the compound that spawned the synthetic chemistry industry in 1856.
Fluorescent probe chemistries
To detect a specific sequence in a PCR product, a synthetic oligonucleotide must assume the role of a molecular device, signalling to the analyst whether it has hybridised or not. In real-time PCR, a fluorescently-labelled oligonucleotide is designed so that it emits a fluorescent signal when hybridised, but remains quenched when in the single-stranded form. In this way, as the PCR proceeds, the fluorescent signal increases as copies of the target sequence are produced. The problem of how to generate a fluorescent signal from a labelled probe has been tackled in several ways, leading to a growth in DNA diagnostic methodologies, including Molecular Beacons and TaqMan probes.4
Molecular Beacons
In this technology a conformational change in the probe following hybridisation generates a fluorescent signal (Fig 2). Molecular Beacons are oligonucleotide probes with a stem-and-loop structure. The probe sequence in the loop portion of the molecule is designed to complement a predetermined target DNA sequence. The stem is formed by two complementary arm sequences, usually four-five bases in length. These are unrelated to the target sequence and are designed to anneal on either side of the probe. A fluorophore is covalently linked to the end of one arm of the stem and a fluorescence-quenching molecule is covalently linked to the end of the other arm. The stem keeps these two moieties in close proximity to each other, causing fluorescence to be quenched when the beacon is in the 'off' position. The quencher, a non-fluorescent chromophore such as DABCYL (5), converts the energy received from the fluorophore into heat.
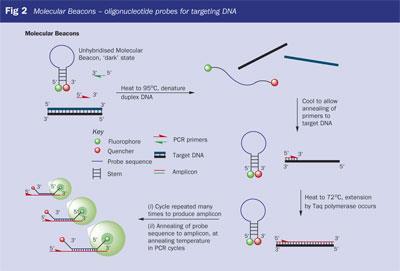
After PCR amplification, when the probe anneals to a target molecule, an intermolecular duplex is formed. This is longer and thermodynamically more stable than the intramolecular closed Molecular Beacon formed by the arm sequences. This occurs only if the PCR product complements the probe sequence of the Molecular Beacon.
The rigidity and the length of the probe-target hybrid preclude the simultaneous existence of the stem-and-loop structure. Consequently, the probe is forced to undergo a conformational change and the arm sequences 'come unstuck' and move away from each other. When this occurs the fluorophore and the quencher are not in close proximity, and the Molecular Beacon becomes fluorescent.
TaqMan probes
The TaqMan assay, the most widely used method for analysing PCR products in real time, is based on detecting a specific PCR product. The latter results from hybridisation of the target sequence to a doubly-labelled fluorogenic probe (the TaqMan probe), which is cleaved enzymatically by Taq polymerase during the amplification reaction (Fig 3). The TaqMan assay utilises fluorescence resonance energy transfer (FRET), a 'through-space' energy transfer phenomenon that has been used extensively as a spectroscopic 'ruler' for examining DNA and protein secondary structure. In the intact TaqMan probe the fluorescent dye and the quencher engage in FRET, so the probe is non-fluorescent or 'dark'.
During PCR, the probe is cleaved (digested) by the inherent 5'-dexonuclease activity of the DNA Taq polymerase, a process which only occurs if and when the TaqMan probe hybridises to the target DNA that is being amplified. As a result, the fluorophore is cleaved from the probe and diffuses away from the quencher. FRET can no longer occur and consequently a fluorescence signal is generated.
The development of real-time PCR has been made possible largely by the introduction of two real-time thermal cyclers with online fluorescence monitoring: the ABI 7700, and the Roche LightCycler. The LightCycler, initially developed by Idaho Technologies, is a carousel instrument that uses thin glass capillaries to contain the PCR reactions with reaction volumes as small as 10μl. Although the sample capacity of the LightCycler (32 capillaries) is less than that of the 7700 (96 wells), the more efficient heat transfer through the thin capillary walls allows the use of rapid cycling conditions. A PCR reaction of 40 cycles can be completed in 15-20 minutes, compensating for the lower sample capacity. In 2002, sales of thermal cyclers in the US and Europe were worth $430 million.4 The miniaturisation and optimisation that will result in the use of real-time PCR to the clinic has been made possible by equipment developed in the US and UK for battlefield applications (detection of biological warfare agents).
Continued development and improvement of these exciting technologies are inevitable, particularly as DNA-based diagnostics become more widely used in medicine. The motivation to reduce cost below the current level of over £10 per test will be a powerful one. Whatever breakthroughs are made, it is certain that chemistry will continue to underpin research into new methods for genetic analysis.
Rohan Ranasinghe is a postdoctoral research fellow, and Tom Brown is professor of bioorganic chemistry in the school of chemistry at the University of Southampton, Highfield, Southampton SO17 1BJ.
Box 1
Introducing genetics
Each cell contains all of an organism's genetic information (or genome), stored as DNA (deoxyribonucleic acid). The long DNA molecule is tightly wound and packaged, together with proteins, as a chromosome. A human cell contains 46 chromosomal DNA molecules - two sets of 23, one inherited from each parent. These DNA molecules can be considered as a set of shorter DNA molecules or genes. There are about 30,000 genes in a set of chromosomes, each of which guide the production of one particular component of a molecule. An organism's genome holds its inherited characteristics, which can be translated by the cellular apparatus to make (express) proteins, which mediate all cellular processes. The mechanism by which this occurs can be described as: 'DNA makes RNA makes proteins' so the sequence of the protein is determined by the sequence of the DNA.
DNA is a linear polymer made up of a nucleotide chain - 2'-deoxyribose-5-phosphate - and a base, either a purine (adenine (A) or guanine (G)) or a pyrimidine (thymine (T) or cytosine (C)). In 1953 chemists Francis Crick and James Watson discovered that DNA occurs in cells as a double helix with two long strands wound around each other, and that the chemical structure of the molecule dictates that A always aligns or pairs with T, and C always pairs with G. It is this base pairing that allows DNA in a cell to copy itself and transfer its information to a new cell. Typically a gene comprises a few thousand base pairs: 2000-3000 for the average bacterium, up to 30,000 for a small plant, and up to 100,000 for the human body.
References
- S. L. Beaucage and M. H. Caruthers, Tetrahedron Lett., 1981, 22, 1859.
- K. B. Mullis, Sci. Am., 1990, 36.
- J. R. Lakowicz, Principles of fluorescence spectroscopy. New York: Plenum, 1983.
- J. Wilhelm and A. Pingoud, Chembiochem., 2003, 4, 1120.









No comments yet