Elinor Hughes discovers the technique that has imaged molecules directly for the first time
Every chemist is familiar with structural drawings of atoms and bonds in textbooks but sometimes it can be difficult to translate those drawings into reality. But what if the drawings were replaced by photographs of molecules showing their real atoms and bonds? Excitingly, scientists have moved a step closer to such photographs with a technique called non-contact atomic force microscopy (ncAFM). The images captured by ncAFM add another dimension to how we learn about bonding in chemical compounds.

The first image of a molecule was taken in 2009 by the atomic manipulation group at IBM Research – Zurich, Switzerland, using an ultra-high vacuum atomic force microscope.1 The molecule was pentacene – five benzene rings fused in a line – and the images created waves in the chemical community.
Then, in 2010, ncAFM was used by the IBM team to assist Marcel Jaspars’ group at the University of Aberdeen, UK. The team at Aberdeen was trying to solve the structure of cephalandole A, a metabolite extracted from the deep-sea bacterium Dermacoccus abyssi. Using the usual method of determining structures – nuclear magnetic resonance spectroscopy, which shows a graph with peaks that represent different atoms in a molecule – they saw they had produced one of four possible structures, but could not narrow it down any further. The ncAFM image showed the correct structure.
Michael Crommie and Felix Fischer and their teams at the University of California, Berkeley, US, went a step further in 2013 and used ncAFM to image the change in the covalent bond structure of a molecule – oligo-(phenylene-1,2-ethynylene) – and its two major products resulting from a cyclisation reaction at the surface.2
The technique
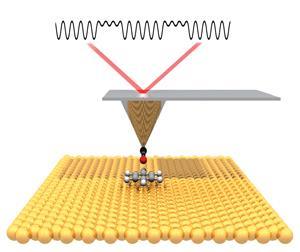
A non-contact atomic force microscope generates high resolution topographic images of materials. To get an image of a molecule, an atomically sharp tip is functionalised with a single carbon monoxide (CO) molecule at its apex. This chemically modified tip is attached to a vibrating quartz tuning fork and is scanned across a surface. The shift in the vibration frequency, caused by forces between the CO tip and the molecule adsorbed to the surface, is monitored by very sensitive electronics and can be represented as a gradient map of the surface.
‘It’s a little bit like reading braille,’ says Felix. ‘You can think about it as moving a finger across a surface and feeling the protrusions on that surface.’ He adds that the tip is oscillated by a quartz crystal, which resonates at such a constant frequency that it is normally used to keep time in most wristwatches. ‘There’s a million dollar instrument around it, but the business end that enables the measurements is a tiny item you can buy in every Radio Shack. It’s such a simple concept that gives you a powerful tool.’
The tip itself can be cut by hand, as teacher Peter Tryon and his A-level chemistry and physics students from a school near Beijing in China discovered when they visited the laboratory of ncAFM imaging expert Xiaohui Qiu at the National Centre for Nanoscience and Technology in Beijing. ‘To produce images with a sufficiently high resolution, the end of the tip will typically be around 10 to 100 nm in diameter,’ says Peter. One of Xiaohui’s PhD students, Pengcheng Chen, demonstrated how he used a microscope, scalpel and tweezers to cut the tip, which is too small to see with the naked eye. When asked how he managed to cut something as thin as 10–100 nm by hand, Pengcheng answered ‘every forest has a tallest tree’, explaining that a sharp tip will naturally have tiny bumps and the one that protrudes most will naturally become the operational tip of the probe.
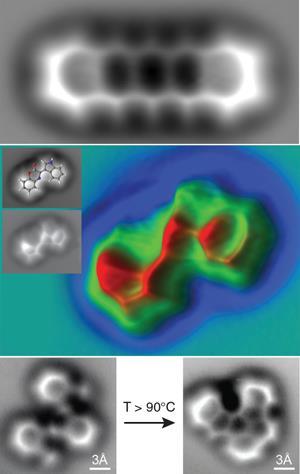
As the tip is metal, there’s a chance it could react with the molecule being analysed. This problem was solved by accident at IBM when Leo Gross and his team unintentionally picked up a CO molecule from the surface on their tip. ‘With CO, the AFM tip can be brought close enough to the molecule on the surface to be able to get atomically resolved images without accidentally moving the molecule around or picking it up by the tip,’ says Peter Liljeroth of Aalto University, Finland, who was working with IBM at the time.
Molecules at room temperature tend to move around, which would not give a clear image, so to image the before and after photos of their cyclisation reaction, Felix and his team cooled the substrate to 4 K. They then condensed the substrate’s molecules onto a surface. As the molecules hit the surface, the team recorded an image, then removed the sample from the AFM stage. They used a simple heating element to heat the substrate to supply enough energy for the molecule to undergo the reaction. ‘We then moved it back to the imaging stage, cooled it to 4 K again and recorded the image,’ says Felix. This was all done under ultra-high vacuum to prevent gas molecules in the air interfering with the results.
Imaging can take a while, depending on the resolution required. ‘Since the tip is moved across the surface pixel by pixel, it depends on how many pixels you want to scan,’ explains Felix. ‘The images in the Science paper took 20–30 minutes.2 The critical factor is that during that time, nobody slams the door to the room or bumps into the instrument, or you have to start again. We once needed a high resolution image that would take around two hours, so we set up the instrument, went home and started it remotely at 3 o’clock in the morning.’
Imaging the hydrogen bond
Unfortunately, the technique has had a small set-back in the area of molecule imaging. In 2013, Xiaohui Qiu and team published images of 8-hydroxyquinoline molecules with what appeared to be lines representing hydrogen bonds between the molecules.3 This promised to be the next big breakthrough until doubt was cast over the origin of the lines in 2014 in a paper published by Pavel Jelínek from the Czech Academy of Science.4 ‘There will be a ridge-like feature that will look like a bond in the images between any two atoms that are close to each other,’ says Peter Liljeroth. His team produced images of molecules that showed these lines between two nitrogen atoms that were close to each other but not bonded.5

Peter explains that when the tip is brought close to the molecule, a repulsion is caused as the electron cloud of the CO overlaps with the electron cloud of the molecule. The CO then bends away a little to reduce the interaction. If the tip is between two atoms, the CO is in a labile position so it could go either way. This causes bright contrast along the line connecting the atoms. ‘It does not necessarily mean there would not be hydrogen bonds in those locations, just that the features in the AFM images can also arise without them,’ he says. He suggests that perhaps in the future, the tip could be functionalised with molecules other than CO to maximise the measuring signal for the type of effect being searched for, including hydrogen bonding.
History of AFM
The atomic force microscope was developed in 1986 by Gerd Binnig, Christoph Gerber and Calvin Quate to overcome the shortcomings of the scanning tunnelling microscope. Unlike ordinary microscopes and electron microscopes, scanning tunnelling microscopy (STM) allowed scientists to visualise objects even smaller than atoms.
Designed in 1981 by Gerd Binnig and Heinrich Rohrer, and later netting them the Nobel prize in physics in 1986, STM provides a 3D atomic scale map of atoms by moving a tip one atom wide across a sample in a vacuum chamber at -271°C. An electric current carried to the tip flows between the tip and the sample and as the tip gets closer to the sample, the current increases. Measuring the current flow gives data about the distance from the tip to the individual atoms, which is translated into an image by a computer. Unfortunately, it can only be used with conductive surfaces such as metals.
To overcome this, the tip of the atomic force microscope was designed to be able to touch the sample so it could be used with non-conductive surfaces. The team attached the tip to a flexible arm – the cantilever – which allowed it to move towards or away from the sample as atomic forces attracted or repelled it. The cantilever’s movements are tracked by a laser and the laser’s angles of reflection are detected by an electronic sensor, which sends the signal to a computer to be translated to an image of the sample’s surface.
Modes of operation include contact mode and non-contact mode. In contact mode, the tip is dragged across a sample’s surface and the surface’s contours are measured using the deflection of the cantilever or using the feedback signal needed to keep the cantilever at a constant position. If the tip gets too close to a surface, strong attractive forces can cause the tip to ‘snap-in’ to the surface. So this type of AFM is done at a depth where the overall force is repulsive, that is, in firm ‘contact’ with the solid surface below any adsorbed layers.
Non-contact mode AFM (ncAFM), unlike other AFM modes, gives true atomic resolution images. The tip does not degrade, making ncAFM more suitable for scanning biological samples and organic thin films. This is because the tip does not come into contact with the sample’s surface, but it can be positioned very close to the sample. This is achieved by oscillating the cantilever at its resonance frequency (frequency modulation) or just above (amplitude modulation) where the amplitude of oscillation is typically a few nanometres (
Challenges
One of the things the molecules imaged so far have in common is that they are all flat, and this is one of the technique’s drawbacks. AFM can’t image anything three-dimensional, so there is a limit to the molecules that can be imaged. ‘If you don’t have a flat surface, you don’t see anything, at least at the level we’re working at,’ says Felix, who points out that the most interesting molecules are three-dimensional, particularly complex natural products and pharmaceutically active compounds. Physicists are working on improving the instrumentation to enable it to image three-dimensional molecules. ‘It’s an engineering problem and I think it’s going to take a few more years before they come up with a solution,’ says Felix.

Another limitation is that AFM can only look at reactions in stages, so it would be difficult to capture images of reaction intermediates, which only exist for a short time. However, physicists such as Peter Liljeroth are working out how to adapt the instrumentation. ‘If you could look at reaction intermediates, this would be very important in terms of understanding how reactions work,’ he says. Progress toward seeing intermediates is already being made.6
The future
It’s difficult to predict the future for ncAFM in imaging molecules as the instrumentation is not universally available and only a handful of companies in the world build the instruments. Until four years ago, there were no commercial instruments available. Even now, they have to be custom built and this can take a year. ‘The capabilities of the instruments keep increasing but imaging molecules is still not something you can do on a routine basis,’ says Felix. ‘At this stage, AFM is not going to replace any of the other analytical tools used as standard.’
Felix suggests the technique could be used to analyse reactions that occur on surfaces, which is difficult to do currently. ‘It’s essentially a black box. You put in the starting material and on the other side pull out a product. You measure the parameters in the black box, but you never really know what’s happening at a molecular level,’ he says. Since most industrial production of chemicals is done on surfaces instead of in solution, gaining information about what happens to a molecule on a surface could help maximise the reaction’s efficiency by tuning the catalyst or environment appropriately.
Since Felix and his team published their paper in Science,2 they have received more than 200 requests for their images from people all over the world. ‘When I studied chemistry, we were told: “you’re never going to be able to see a single molecule”. That was just 10 years ago,’ says Felix. ‘The closest they got was a crystal structure, which is an averaged image over millions of molecules. Now, scientists are able to look at a single molecule with almost the same or even better resolution. The tool is inspiring. I wish we had had textbooks with these images. Pupils can now start to see a real perspective; it’s not something abstract anymore.’
Elinor Hughes is a science writer based in Bridgend, UK
References
1. L Gross et al, Science, 2009, 325, 1110 (DOI: 10.1126/science.1176210)
2. D G de Oteyza et al, Science, 2013, 340, 1434 (DOI: 10.1126/science.1238187)
3. J Zhang et al, Science, 2013, 342, 611 (DOI: 10.1126/science.1242603)
4. P Hapala et al, Phys. Rev. B, 2014, 90, 085421 (DOI: 10.1103/PhysRevB.90.085421)
5. S K Hämäläinen et al, Phys. Rev. Lett., 2014, 113, 186102 (DOI: 10.1103/PhysRevLett.113.186102)
6. Chemistry World, September 2015









No comments yet