- Home
- I am a …
- Resources
- Collections
- Remote teaching support
- Starters for ten
- Screen experiments
- Assessment for learning
- Microscale chemistry
- Faces of chemistry
- Classic chemistry experiments
- Nuffield practical collection
- Anecdotes for chemistry teachers
- Literacy in science teaching
- More …
- Climate change and sustainability
- Alchemy
- On this day in chemistry
- Global experiments
- PhET interactive simulations
- Chemistry vignettes
- Context and problem based learning
- Journal of the month
- Chemistry and art
- Classic chemistry demonstrations
- In search of solutions
- In search of more solutions
- Creative problem-solving in chemistry
- Solar spark
- Chemistry for non-specialists
- Health and safety in higher education
- Analytical chemistry introductions
- Exhibition chemistry
- Introductory maths for higher education
- Commercial skills for chemists
- Kitchen chemistry
- Journals how to guides
- Chemistry in health
- Chemistry in sport
- Chemistry in your cupboard
- Chocolate chemistry
- Adnoddau addysgu cemeg Cymraeg
- The chemistry of fireworks
- Festive chemistry
- Collections
- Education in Chemistry
- Teach Chemistry
- Events
- Teacher PD
- Enrichment
- Our work
- More navigation items
Close menu
- Home
- I am a …
-
Resources
- Back to parent navigation item
- Resources
- Primary
- Secondary
- Higher education
- Curriculum support
- Practical
- Analysis
- Literacy in science teaching
- Periodic table
- Climate change and sustainability
- Careers
- Resources shop
-
Collections
- Back to parent navigation item
- Collections
- Remote teaching support
- Starters for ten
- Screen experiments
- Assessment for learning
- Microscale chemistry
- Faces of chemistry
- Classic chemistry experiments
- Nuffield practical collection
- Anecdotes for chemistry teachers
- Literacy in science teaching
- More …
- Climate change and sustainability
- Alchemy
- On this day in chemistry
- Global experiments
- PhET interactive simulations
- Chemistry vignettes
- Context and problem based learning
- Journal of the month
-
Chemistry and art
- Back to parent navigation item
- Chemistry and art
- Techniques
- Art analysis
- Pigments and colours
- Ancient art: today's technology
- Psychology and art theory
- Art and archaeology
- Artists as chemists
- The physics of restoration and conservation
- Cave art
- Ancient Egyptian art
- Ancient Greek art
- Ancient Roman art
- Classic chemistry demonstrations
- In search of solutions
- In search of more solutions
- Creative problem-solving in chemistry
- Solar spark
- Chemistry for non-specialists
- Health and safety in higher education
- Analytical chemistry introductions
- Exhibition chemistry
- Introductory maths for higher education
- Commercial skills for chemists
- Kitchen chemistry
- Journals how to guides
- Chemistry in health
- Chemistry in sport
- Chemistry in your cupboard
- Chocolate chemistry
- Adnoddau addysgu cemeg Cymraeg
- The chemistry of fireworks
- Festive chemistry
- Education in Chemistry
- Teach Chemistry
- Events
- Teacher PD
- Enrichment
- Our work
Journal articles made easy: Predict crystallinity
2015-07-14T13:28:56
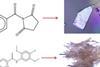
This article looks at predicting and controlling the crystallinity of molecular materials. It will help you understand the research the journal article is based on, and how to read and understand journal articles. The research article was originally published in our CrystEngComm journal.
Make your lessons pop
Choose an account option to continue exploring our full range of articles and teaching resources
Already have an account?
Sign in now
Free
Free access for everyone, everywhere. If you only need a few resources, start here.
What's included
- One free teaching resource each month
- Five free Education in Chemistry articles each month
- Personalised email alerts and bookmarks
UK and Ireland only
Free
for eligible users
Free and comprehensive access for teachers and technicians at secondary schools, colleges and teacher training institutions in the UK and Ireland.
What's included
- Unlimited access to our resources and practical videos
- Unlimited access to Education in Chemistry articles
- Access to our online assessments
- Our teacher well-being toolkit and personal development resources
- Applications for funding to support your lessons
£80
per year
Get unlimited articles and resources each month, plus discounts on professional development courses.
What's included
- Unlimited access to our resources and practical videos
- Unlimited access to Education in Chemistry articles
- 10% off our self-led professional development courses
Already have an account? Sign in now